Gene Set Enrichment Analysis (GSEA) with fgsea - easy R tutorial
HTML-код
- Опубликовано: 9 сен 2023
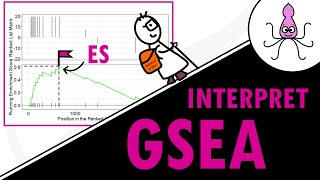
- In this tutorial, I will explain how to perform gene set enrichment analysis on your differential gene expression analysis results. We will use the R package fgsea() and you will learn how to:
- Install and start fgsea()
- Prepare your dataset to perform GSEA
- Set the analysis parameters and run the analysis.
- View the GSEA results and get some nice plots!
And as always, you can find the code I am using in this tutorial at biostatsquid.com, where you can also find a step by step explanation of the code. For this tutorial you will need R, or Rstudio, and you will need to install the fgsea() package.
Hope you like it!
biostatsquid.com/fgsea-tutori...
--------------------------------------------------------------------------------------------------------------------
Watched it already?
If you liked this video or found it useful, please let me know! Your comments and feedback are very much appreciated😊
If you have questions, don't hesitate to leave me a comment down below, I will answer as soon as I can:)
--------------------------------------------------------------------------------------------------------------------
Are you into biostatistics and computational analysis?
For more biostatistics tools and resources, you can visit:
biostatsquid.com/
Follow me on Instagram at @biostatsquid:
/ biostatsquid
For more
• simple and clear explanations of biostatistics methods
• computational biology tools
• easy step-by-step tutorials in R and Python
to analyse and visualise your biological data!
Don’t forget to subscribe if you don’t want to miss another video from me! --------------------------------------------------------------------------------------------------------------------
Other interesting resources for pathway enrichment analysis:
Other interesting resources for GSEA:
Main GSEA webpage: www.gsea-msigdb.org/gsea/inde...
More on the method itself: www.pnas.org/doi/10.1073/pnas...
Paper comparing pathway enrichment analysis methods: www.nature.com/articles/s4159...
Music from #Uppbeat (free for Creators!):
uppbeat.io/t/pecan-pie/east-s...
License code: 80ZOLNQ8NSDSWKFV








I am with zero experience, and failed so many times by following youtubers, you script works and I can easily catch up, even different methods. Thankyou sooooooooomuch.
Another great video! Thank you Biostatsquid, see ya in the next one!
Great concise tutorial!
Much appreciated!👍
Excellent tutorial, thank you!
Many thanks for this video. It was extremely helpful! Just a quick question, do you have a link to any papers that use the same method for ranking genes? I've gone for the same approach, but will need to defend it in my viva and I am struggling to find publications using this method.
Secondly, I just want to confirm that you use regular p-values rather than adjusted p-values for the ranking calculation?
Perhaps you didn't know, but now there's a software platform called RNAlysis that is highly modular and contains a graphical user interface so that you can interrogate any RNA based research question and never have to write a single line of code.
A prebuilt software will never have the flexibility of coding. Said this, maybe for entry level is just fine... but in real research you may need to explore different settings to get the best results.
Can you please put your all your code?
Hi! Yes of course, you can find it here: biostatsquid.com/fgsea-tutorial-gsea/
Promo`SM 😳
Thank you for the nice tutorial, I have two questions:
1/ From the Github-page of fgsea (issue #131 'Which genes should I use"), the developer seems to mention that fgsea automatically extracts background genes from the sorted input vector. You use this custom function to filter the pathways yourself. I ran both options (i.e. once without filtering and once with filtering) and results are very comparable, but NES-values and P-values are not exactly the same. Would you still recommend using the function?
2/ To rank genes, you use (df$log2FC)*(-log10(df$PValue)). Do you have any references to the use of this formula? One of the developers advises to use the test-statistic to rank (i.e. t column from limma and stat column from DESeq2). I have also seen ranking be done based on log2FC.
Why do you advise this formula?
Thank you.
Hi, thanks so much for your comment, two very good questions.
1. I wasn't aware that fgsea already filters out genes from the gene sets - not sure if the comment you referred to means that, I would need to check the methods or the fgsea funtion itself to confirm this. But thanks so much for asking and letting me know! Once I check it I'll post it:)
In case you want to read about this filtering, this recent article covers the topic. genomebiology.biomedcentral.com/articles/10.1186/s13059-015-0761-7
2. You're totally on point, I've also seen different methods. I guess I'd rather go for a combination of both because I want to rank them also by significance, not only fold change. But again, depends on your project and what you're looking for. I think I shared a paper already on my blogpost which recommends this ranking method - www.ncbi.nlm.nih.gov/pmc/articles/PMC3957066/